Description
The Topology ToolKit (TTK) is an open-source library and software collection for topological data analysis in scientific visualization.
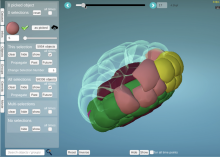
TTK can handle scalar data defined either on regular grids or triangulations, either in 2D or in 3D. It provides a substantial collection of generic, efficient and robust implementations of key algorithms in topological data analysis. It includes:
· For scalar data: critical points, integral lines, persistence diagrams, persistence curves, merge trees, contour trees, Morse-Smale complexes, topological simplification;
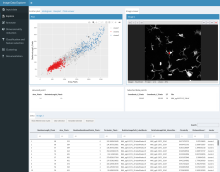
· For bivariate scalar data: fibers, fiber surfaces, continuous scatterplots, Jacobi sets, Reeb spaces;
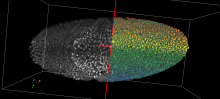
· For uncertain scalar data: mandatory critical points;
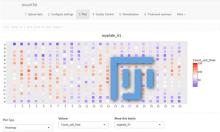
· For time-varying scalar data: critical point tracking;
· For high-dimensional / point cloud data: dimension reduction;
· and more!
TTK makes topological data analysis accessible to end users thanks to easy-to-use plugins for the visualization front end ParaView. Thanks to ParaView, TTK supports a variety of input data formats.
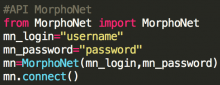
TTK is written in C++ but comes with a variety of bindings (VTK/C++, Python) and standalone command-line programs. It is modular and easy to extend. We have specifically developed it such that you can easily write your own data analysis tools as TTK modules.