Description
Deep learning based image restoration methods have recently been made available to restore images from under-exposed imaging conditions, increase spatio-temporal resolution (CARE) or self-supervised image denoising (Noise2Void). These powerful methods outperform conventional state-of-the-art methods and leverage down-stream analyses significantly such as segmentation and quantification.
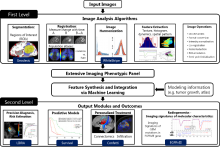
To bring these new tools to a broader platform in the image analysis community, we developed a simple Jupyter based graphical user interface for CARE and Noise2Void, which lowers the burden for non-programmers and biologists to access these powerful methods in their daily routine. CARE-less supports temporal, multi-channel image and volumetric data and many file formats by using the bioformats library. The user is guided through the different computation steps via inline documentation. For standard use cases, the graphical user interface exposes the most relevant parameters such as patch size and number of training iterations, while expert users still have access to advanced parameters such as U-net depth and kernel sizes. In addition, CARE-less provides visual outputs for training convergence and restoration quality. Any project settings can be stored and reused from command line for processing on compute clusters. The generated output files preserve important meta-data such as pixel sizes, axial spacing and time intervals.