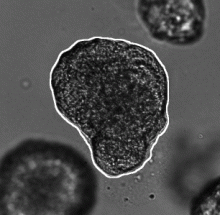
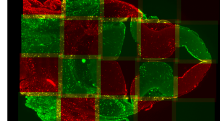
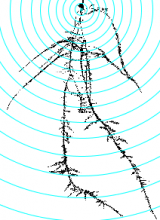
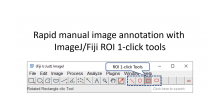
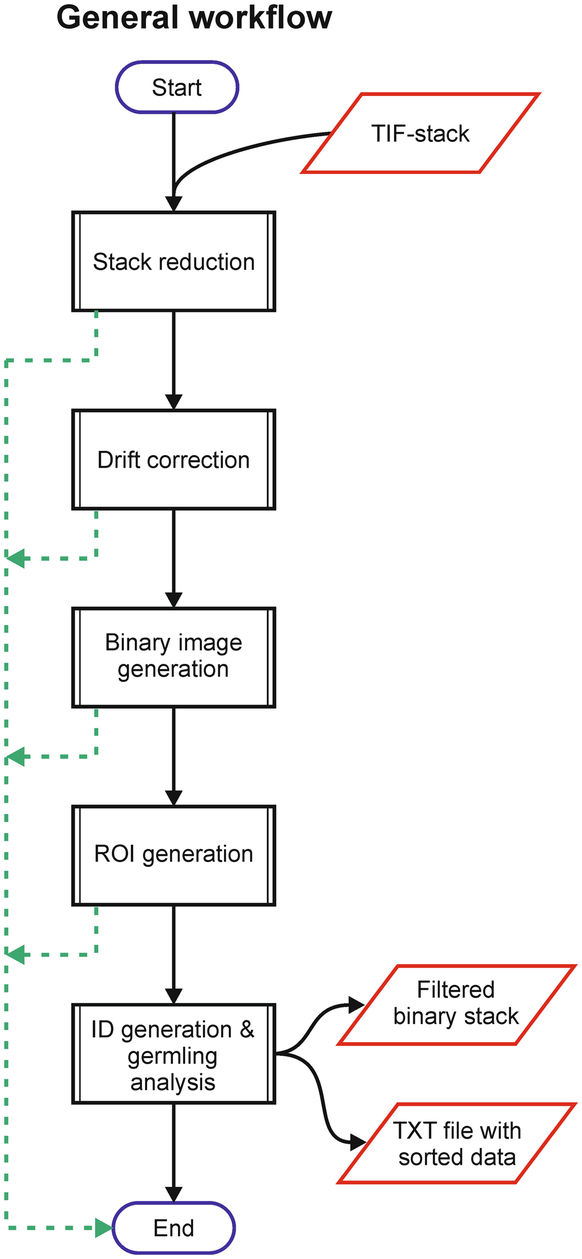
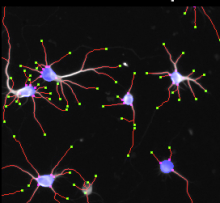
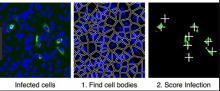
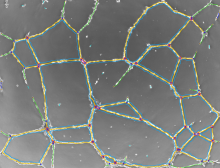
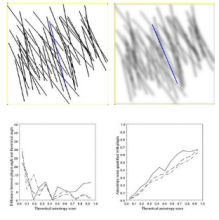
GPU-Accelerating ImageJ Macro Image Processing Workflows Using CLIJ
This chapter is part of this book. The chapter introduces GPU-accelerated image processing in ImageJ/Fiji. The reader is expected to have some pre-existing knowledge of ImageJ Macro programming. Core concepts such as variables, for-loops, and functions are essential. The chapter provides basic guidelines for improved performance in typical image processing workflows.