Description
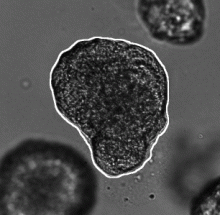
This workflow describes a deep-learning based pipeline for reliable single-organoid segmentation and tracking in 2D+t high-resolution brightfield microscopy of mouse mammary epithelial organoids. The pipeline involves a four-layer U-Net to infer semantic segmentation predictions, adaptive morphological filtering to establish candidate organoid instances, and a shape-similarity-constrained, instance-segmentation-correcting tracking step to associate the corresponding organoid instances in time.
It is particularly focused on automatically detecting an organoid located approximately in the center of the first frame and track all its subsequent instances in the remaining frames, emphasizing on accurate organoid boundary delineation. Furthermore, segmentation network was trained using plausible pix2pixHD-generated bioimage data. Syntheric image simulator code and data are also available here.