Human colon tissue
- Read more about Human colon tissue
- 193 views
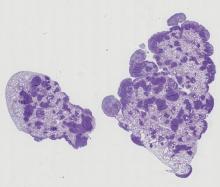
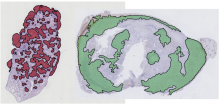
One of the principal challenges in counting or segmenting cells or cell nuclei is dealing with clustered objects such as in tissues. To help assess algorithms' performance in this regard, synthetic 3D image sets of human colon tissue are provided in two diferent levels of quality: high SNR and low SNR. Ground truth is available as well.