Description
Various ways are proposed in different websites for example:
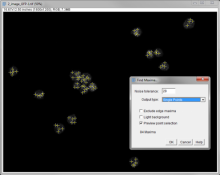
Here, a workflow template using ImageJ's build-in Find Maxima ( Process -> Find Maxima) is explained. It can be used for many 2D counting-related tasks.
For counting small, bright foci (dots), set Output type to be Point Selection. If too many points are detected, the number may be reduced using one or more of the following methods:
Apply a filter to reduce noise, e.g. Process -> Filters -> Gaussian Blur... prior to running Find Maxima
Set a minimum threshold with Image -> Adjust -> Threshold... prior to running Find Maxima, then use the Above lower threshold option within the dialog box
Increase the Noise tolerance value (which effectively acts as a local threshold)
The resulting point selection can be modified (points added/removed) by the Multi-Point tool.
After the points are available, final measurements can be made using Analyze -> Measure.