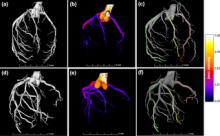
Workflow
A workflow is a set of components assembled in some specific order to
- Measure and estimate some numerical parameters of the biological system or
- Visualization
for addressing a biological question. Workflows can be a combination of components from the same or different software packages using several scripts and manual steps.