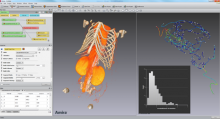
Image registration
Image registration is the process of transforming different sets of data into one coordinate system. Registration is necessary in order to be able to compare or integrate the data obtained from different sensors/imaging modalities, at different times, from different view points, etc. . Registration can be based on correspondence established between the landmarks or feature points. Alternatively, some similarity/distance metric is established between the image intensity maps to navigate the registration process.
Synonyms
Image alignment