The Allen Cell Structure Segmenter
Type
Requires
Implementation Type
Programming Language
Supported image dimension
Interaction Level
License/Openness
License
2-clause BSD license plus clause a third clause that prohibits redistribution for commercial purposes without further permission.
Description
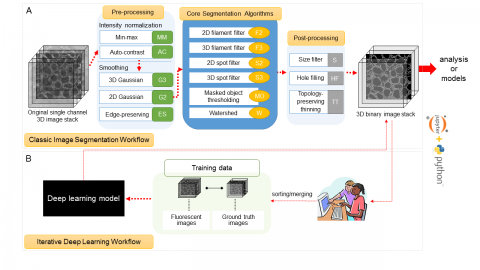
The Allen Cell Structure Segmenter is a Python-based open source toolkit developed at the Allen Institute for Cell Science for 3D segmentation of intracellular structures in fluorescence microscope images.
It consists of two complementary elements:
- Classic image segmentation workflows for 20 distinct intracellular structure localization patterns. A visual “lookup table” is outlining the modular algorithmic steps for each segmentation workflow. This provides an intuitive guide for selection or construction of new segmentation workflows for a user’s particular segmentation task.
- Human-in-the-loop iterative deep learning segmentation workflow trained on ground truth manually curated data from the images segmented with the segmentation workflow. Importantly, this module was not released yet.
has function
has topic
has biological terms
Additional keywords
Post date
01/02/2019 - 10:09
Last modified
01/02/2019 - 18:15