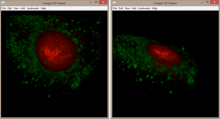
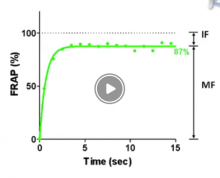
Description
OMERO is an image database application consisting of a server and several clients, the most important of which are the web client and _Insight_ java application. Metadata are extracted from images that have been imported (either using the Insight client, or directly from the filesystem), and this is accessible for search. A standardised hierarchy of _Project > Dataset > Image_ in which image thumbnails can be viewed, combined with group membership, tagging, and attachment of results and other files gives a powerful framework for organising scientific image data. Images can also be analysed server-side or client-side within the base OMERO application or one of its many extensions. OMERO has APIs for extension in multiple languages: java, python, C++ and MATLAB; and such extensions have easy access to the image data and metadata in the database.