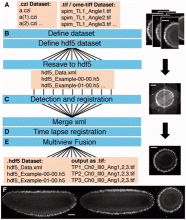
EB1 tracking with Matlab
This module follow EB1 tracking with IJ. In this session, we will visualize the tracking results and also cover typical analysis protocols for the quantitative analysis of movement. Two dynamic numerical features could be extracted from tracking results: speed and direction. Estimation of movement speed from multiple trajectories is a popular indicator of movement, and we will quickly go over the method for estimating the average speed of EB1 movement along microtubule. Movement direction is another quantitative feature, but is rarely explored.