Contents
| Image | Title | Category | Type | Description | Updated |
|---|---|---|---|---|---|

|
InfectionCounter | Software | Workflow | Estimate the frequency of hepatitis C virus infected cells based on the intensity of viral antigen associated immunofluorescence. The core is an ImageJ Macro, so it's easy to modify for one's own needs (Link to the code). |
05/16/2018 - 22:40 |
|
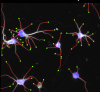
NeurphologyJ | Software | Workflow | ImageJ macro for the morphometry of neurites. > NeurphologyJ; it is capable of automatically quantifying neuronal morphology such as soma number and size, neurite length, neurite ending points and attachment points. NeurphologyJ is implemented as a plugin to ImageJ, an open-source Java-based image-processing and analysis platform.
|
04/27/2018 - 02:14 |

|
Analyze Particles | Software | Component | An object detection function in ImageJ. [Analyze > Analyze Particles...]. It could simply be used for counting number of cells, but could also do more complex stuffs. ## Jython Snippet Here is a snippet of how to use Particle Analysis in Jython script. |
04/25/2023 - 19:55 |

|
Voronoi ImageJ | Software | Component | A resident function in ImageJ, located in the menu as [Process > Binary > Voronoi]. Quote from the ImageJ reference page: |
04/29/2023 - 11:43 |

|
ViBE-Z | Software | Collection | The Virtual Brain Explorer (ViBE-Z) is a software that automatically maps gene expression data with cellular resolution to a 3D standard larval zebrafish (Danio rerio) brain. It automatically detects 14 predefined anatomical landmarks for aligning data. It also offers a database and atlas. The ViBE-Z database, atlas and software are provided via a web interface. A data preparation step is needed in order to provide the right input data and format. |
05/19/2021 - 20:55 |

|
Auto-Bayes | Software | Collection | Auto-Bayes is a software package based on Bayesian statistics and requires no user dependent parameters for molecule detection and image reconstruction for Single-Molecule Localization Microscopy (SMLM), including photoactivated localization microscope (PALM), stochastic optical reconstruction microscope (STORM), and direct stochastic optical reconstruction microscopy (dSTORM), etc. |
03/05/2020 - 12:59 |

|
SynPAnal | Software | Collection | This software is designed for the rapid semi-automatic detection and quantification of synaptic protein puncta from 2D immunofluorescence images generated by confocal laser scanning microscopy. |
03/05/2020 - 13:06 |

|
StabiTissue | Software | Component | - 2D Stabilization in each slice of the stacks in time. - 3D Stabilization intravital imaging of all the stacks (including the dimension Z) - create the videos and the stabilized images in a new folder 2701 |
10/21/2019 - 10:39 |
|
PSF Lab | Software | Component | PSF Lab is a software program that calculates the illumination point spread function of a confocal microscope under various imaging conditions. It is available in 32-bit and 64-bit for Windows and in 64-bit for Mac. |
10/16/2019 - 18:47 |

|
FOCAL | Software | Collection | FOCAL (Fast Optimized Cluster Algorithm for Localizations) is a rapid density based algorithm for detecting clusters in localization microscopy datasets. |
09/13/2017 - 15:22 |

|
MIATool | Software | Collection | The Microscopy Image Analysis Tool (MIATool) is a software application designed for the viewing and processing of N-dimensional array of images. At its core is an image viewer which allows the traversal of an N-dimensional array of images. Besides the standard display as pixels of varying intensity values, options are available to view the images as mesh or contour plots. The current version of MIATool supports four different image editing tools which can be used to process the images displayed in the viewer. | 09/13/2017 - 12:16 |

|
WaveTracer | Software | Component | WaveTracer is a plugin for Metamorph. It represents a functional method for real-time reconstruction with automatic feedback control, without compromising the localization accuracy. It relies on a wavelet segmentation algorithm, together with a mix of CPU/GPU implementation. |
10/18/2019 - 19:10 |

|
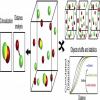
Clus-Doc | Software | Collection | Clus-Doc is a software that quantifies both the spatial distribution of a protein as well as its colocalization status. It may be used to quantify signaling activity and protein colocalization at the level of individual proteins. |
09/13/2017 - 15:17 |
|
Distance Analysis (DiAna) | Software | Component | This plugin allows : Calculating co-localization between objects in 3D Measuring 3D distances between nearest object, co-localized or not Getting some 3D measurements about each objects The plugin can be used with labelled images, but it also integrates tools for the segmentation of the objects. Programming language: JAVA Processes: Denoise filter Segmentation of the objects Object based co-localization and distance analysis Counting and measurements on objects |
05/03/2023 - 17:17 |

|
2D-3D distributed parallel region competition segmentation | Software | Component | This is the source code and data page of a distributed parallel algorithm 2683 for segmentation of large fluorescence microscopy images. |
10/18/2019 - 19:07 |
|
MosaicSuite | Software | Collection | Image-processing algorithms developed at the MOSAIC Group for fluorescence microscopy. Tools included: |
05/08/2023 - 03:12 |

|
Balanced SOFI | Software | Collection | Super-resolution optical fluctuation imaging (SOFI) achieves 3D super-resolution by computing temporal cumulants or spatio-temporal cross-cumulants of stochastically blinking fluorophores. In contrast to localization microscopy, SOFI is compatible with weakly emitting fluorophores and a wider range of blinking conditions. Balanced SOFI analyses several cumulant orders for extracting molecular parameter maps, such as the bright and dark state lifetimes, the concentration and the brightness distributions of fluorophores within biological samples. |
05/03/2023 - 11:51 |

|
Morpholeaf | Software | Component | The MorphoLeaf application allows you to extract the contour of multiple leaf images and identify their biologically-relevant landmarks. These landmarks are then used to quantify morphological parameters of individual leaves and to reconstruct average leaf shapes. MorphoLeaf is developed by the Modeling and Digital Imaging and the Transcription Factors and Architecture teams of the Institut Jean-Pierre Bourgin, INRA Versailles, France, and the Biophyscis and Development group at RDP, Lyon. |
10/18/2018 - 17:31 |
| |
FreeD | Software | Collection | Free-D is a three-dimensional (3D) reconstruction and modeling software. It allows to generate, process and analyze 3D point and surface models from stacks of 2D images. Free-D is an integrated software, offering in a single graphical user interface all the functionalities required for 3D modeling. It runs on Linux, Windows, and MacOS. Free-D is developed by the Modeling and Digital Imaging team of the Institut Jean-Pierre Bourgin, INRA Versailles, France. |
10/18/2018 - 17:39 |

|
SuReSim | Software | Collection | SuReSim (Super Resolution Simulation) is an open-source simulation software for Single Molecule Localization Microscopy (SMLM). The workflow of the SuReSim algorithm starts from a ground truth structure and lets the user choose to either directly simulate 3D localizations or to create simulated *.tiff-stacks that the user can analyze with any given SMLM reconstruction software. A 3D structure of any geometry, either taken from electron microscopy, designed de-novo from assumptions or known structural facts, is fluorophore-labeled in silico. |
10/19/2018 - 12:43 |
