Description
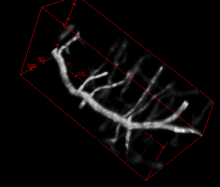
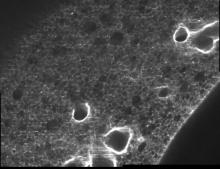
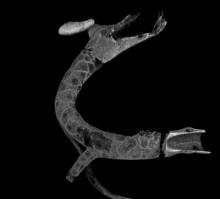
Marker-controlled Watershed is an ImageJ/Fiji plugin to segment grayscale images of any type (8, 16 and 32-bit) in 2D and 3D based on the marker-controlled watershed algorithm (Meyer and Beucher, 1990). This algorithm considers the input image as a topographic surface (where higher pixel values mean higher altitude) and simulates its flooding from specific seed points or markers. A common choice for the markers are the local minima of the gradient of the image, but the method works on any specific marker, either selected manually by the user or determined automatically by another algorithm. Marker-controlled Watershed needs at least two images to run: The Input image: a 2D or 3D grayscale image to flood, usually the gradient of an image. The Marker image: an image of the same dimensions as the input containing the seed points or markers as connected regions of voxels, each of them with a different label. They correspond usually to the local minima of the input image, but they can be set arbitrarily. And it can optionally admit a third image: The Mask image: a binary image of the same dimensions as input and marker which can be used to restrict the areas of application of the algorithm. Set to "None" to run the method on the whole input image. Rest of parameters: Calculate dams: select to enable the calculation of watershed lines. Use diagonal connectivity: select to allow the flooding in diagonal directions.