- 274 views
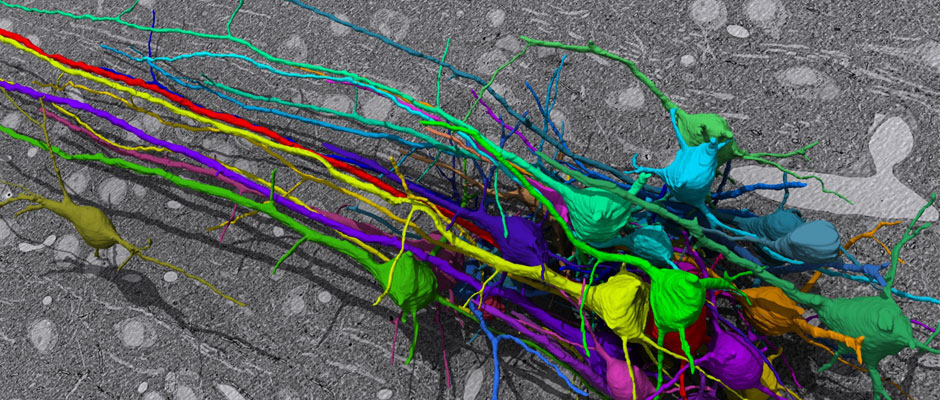
In this challenge, a full stack of electron microscopy (EM) slices will be used to train machine-learning algorithms for the purpose of automatic segmentation of neurites in 3D. This imaging technique visualizes the resulting volumes in a highly anisotropic way, i.e., the x- and y-directions have a high resolution, whereas the z-direction has a low resolution, primarily dependent on the precision of serial cutting. EM produces the images as a projection of the whole section, so some of the neural membranes that are not orthogonal to a cutting plane can appear very blurred. None of these problems led to major difficulties in the manual labeling of each neurite in the image stack by an expert human neuro-anatomist.
The image data used in the challenge was produced by Lichtman Lab at Harvard University (Daniel R. Berger, Richard Schalek, Narayanan "Bobby" Kasthuri, Juan-Carlos Tapia, Kenneth Hayworth, Jeff W. Lichtman) and manually annotated by Daniel R. Berger. Their corresponding biological findings were published in Cell (2015).
All labels (2D membrane probabilities and 3D labels) for 3D segmentation are provided for one data set, the other data sets has not known ground truth since it is used for the competition.
Volume EM