Contents
| Image | Title | Category | Type | Description | Updated |
|---|---|---|---|---|---|

|
Effective image visualization for publications – a workflow using open access tools and concepts | Software | Workflow | Today, 25% of figures in biomedical publications contain images of various types, e.g. photos, light or electron microscopy images, x-rays, or even sketches or drawings. Despite being widely used, published images may be ineffective or illegible since details are not visible, information is missing or they have been inappropriately processed. The vast majority of such imperfect images can be attributed to the lack of experience of the authors as undergraduate and graduate curricula lack courses on image acquisition, ethical processing, and visualization. |
05/24/2023 - 14:07 |
|
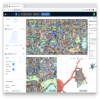
webKnossos: 3D image annotation, visualization and sharing | Software | Collection | webKnossos is an open-source data sharing and annotation platform for tera-scale 2D and 3D image datasets. The core features of webKnossos are: |
03/08/2021 - 14:00 |
|
Viv | Software | Component | Viv is a JavaScript library providing utilities for rendering primary imaging data. Viv supports WebGL-based multi-channel rendering of both pyramidal and non-pyramidal images. The rendering components of Viv are provided as Deck.gl layers, facilitating image composition with existing layers and updating rendering properties within a reactive paradigm. |
02/05/2021 - 00:58 |

|
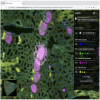
BTrackMate | Software | Component, Workflow | TrackMate based tracker to be used when uploading integer labelled segmentation images, coming from a Deep Learning tool such as stardist. To use this tool efficiently we provide a python notebook to collect/localize the position of cells, this step creates a CSV file which can then be loaded into the Fiji tracker to do particle tracking with TrackMate interface. |
01/28/2021 - 14:50 |
|
A simple MSER based segmentation tool for spot detection in 2/3/4D. | Software | Component | MSER based on implementation in imglib2 provided as an interactive GUI tool for spot detection in 2/3/4D images. |
01/25/2021 - 01:16 |

|
LaRoME | Software | Component | 01/23/2021 - 04:20 | |
|
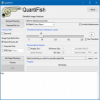
QuantiFish | Software | Workflow | QuantiFish is a quantification program intended for measuring fluorescence in images of zebrafish, although use with images of other specimens is possible. This package is geared towards analysis of fluorescent infection models. The software is designed to automate processing of images of single fish, and outputs results as a .csv file. Alongside measures of total fluorescence above a threshold, this package also introduces several measures for dissemination and distribution of fluorescence throughout the specimen. |
01/06/2021 - 19:51 |
| Data Science in Cell Imaging (DSCI) course material | Training Material | Graduate course at the department of Software and Information Systems Engineering at Ben-Gurion University of the Negev. The recent explosion in high-content, dynamic and multidimensional imaging data is transforming cell imaging into a “Data Science” field. This course will review the state-of-the-art in visualizing, processing, integrating and mining massive cell image data sets, deciphering complex patterns and turning them into new biological insight. It will include a mix of approaches in computer science, machine learning and computer vision applied to bio-imaging data. |
12/31/2020 - 10:15 | ||

|
HistoCAT | Software | Collection | Histology Topography Cytometry Analysis Toolbox (histoCAT) is a package to visualize and analyse multiplexed image cytometry data interactively. It can also export data in.fcs data for further analysis using a specialized cytometry sofwtare such as Flowjo. It can be run as a compiled standalone or from matlab. |
12/09/2020 - 14:52 |

|
NDPItools | Software | Component | Using a Hamamatsu slide scanner such as the NanoZoomer, you may end up with NDPI files that can't always be directly open in standard image analysis software such as ImageJ. |
12/07/2020 - 20:03 |

|
ndpi_extractroi | Software | Component | This small plugin demonstrates the use of OpenSlide in java: it will extract an imageJ roi drawn from the thumbnail of the whole slide image, or the full image at the desired resolution from an hammatsu NDPI file. Note that z stack are not supported by openslide (neitheir ndpiS). |
12/07/2020 - 19:55 |
| |
Image-classification with deep learning in KNIME | Software | Workflow | Set of KNIME workflows for the training of a deep learning model for image-classification with custom images and classes. The workflows take ground-truth category annotations as a table generated by the qualitative annotations plugins in Fiji. Workflows for the training of a model AND for the prediction of image-category for new images are provided. There are different workflows if you do: |
11/20/2020 - 11:09 |
| |
Qualitative annotations Fiji plugins | Software | Component | Set of Fiji plugins facilitating the systematic manual annotation of images or image-regions. From a list of user-defined keywords, these plugins generate an easy-to-use graphical interface with buttons or checkboxes for the assignment of single or multiple pre-defined categories to full images or individual regions of interest. In addition to qualitative annotations, any quantitative measurement from the standard Fiji options can also be automatically reported. |
03/05/2021 - 11:51 |

|
3D Nuclei Clustering Tool | Software | Workflow | Analyze the clustering behavior of nuclei in 3D images. The centers of the nuclei are detected. The nuclei are filtered by the presence of a signal in a different channel. The clustering is done with the density based algorithm DBSCAN. The nearest neighbor distances between all nuclei and those outside and inside of the clusters are calculated. |
05/24/2023 - 16:22 |

|
Interactive features visualization in KNIME | Software | Component | KNIME workflow to visualize a dataset described by multiple quantitative features (ex: a list of samples or cells, each described with multiple morphological features) as a 3D cloud of points (each point corresponding to one sample/cell) as well as a line plot (1 line per sample/cell). |
10/14/2020 - 09:40 |

|
MoBIE-Utils-Python | Software | Collection, Component | The library contains several helper functions to generate MoBIE project folders and add data to it. Itis a python library to generate data in the MoBIE data storage layout. For further information, look to http://biii.eu/mobie-fiji-viewer |
09/09/2020 - 13:43 |
| |
MoBIE Fiji Viewer | Software | Component | MoBIE (Multimodal Big Image Data Exploration) is a framework for sharing and interactive browsing of multimodal big image data. The MoBIE Fiji viewer is based on BigDataViewer and enables browsing of MoBIE datasets. It is also called Platybrowser, and uses the n5 format. |
03/06/2023 - 16:32 |

|
Systems Science of Biological Dynamics database (SSBD:database) | Dataset | This database provides a rich set of open resources for analyzing quantitative data and microscopy images of biological objects, such as single-molecule, cell, gene expression nuclei, etc. Quantitative biological data and microscopy image are collected from a variety of species, sources and methods. These include data obtained from both experiment and computational simulation. Images are downloadable as uploaded raw files. SSBD utilises OMERO internally to manage microscopic images and their metadata. |
09/09/2020 - 15:06 | |
| Bioimage analysis with Icy | Training Material | Short description This webinar is part the serie of webinars from the NEUBIAS Academy @home Recording: https://www.youtube.com/watch?v=5SJqhKjCbMQ&feature=youtu.be . Slides: https://marionlouveaux.github.io/bioimage_analysis_with_icy/ |
05/12/2020 - 17:08 | ||
| |
nd-safir | Software | Component | ND-SAFIR is a software for denoising n-dimentionnal images especially dedicated to microscopy image sequence analysis. It is able to deal with 2D, 3D, 2D+time, 3D+time images have one or more color channel. It is adapted to Gaussian and Poisson-Gaussian noise which are usually encountered in photonic imaging. Several papers describe the detail of the method used in ndsafir to recover noise free images (see references). It is available either in Metamorph (commercial version), either as command line tool. Source are available on demand. |
10/19/2020 - 16:47 |
