Focal Adhesion Analysis Server
The website implements a set of computer vision algorithms designed to automatically process time-lapse images of fluorescently labeled focal adhesion proteins in motile cells.
The methods associated with the processing have been published in PLOS One and Cell. The publication describes a quantitative analysis of focal adhesion dynamics that have been imaged using TIRF. All image processing steps are well explained or referenced.
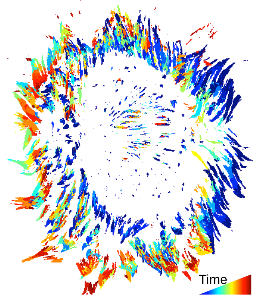
To better understand the dynamic regulation of focal adhesions, we have developed an analysis system for the automated detection, tracking, and data extraction of these structures in living cells. This analysis system was used to quantify the dynamics of fluorescently tagged Paxillin and FAK in NIH 3T3 fibroblasts followed via Total Internal Reflection Fluorescence Microscopy (TIRF). High content time series included the size, shape, intensity, and position of every adhesion present in a living cell. These properties were followed over time, revealing adhesion lifetime and turnover rates, and segregation of properties into distinct zones.