Documentation: Roles, Entry standard and how to curate (read first)
Documentation: Roles, Entry standard and how to curate (read first) lascholz Mon, 02/04/2019 - 18:30Roles
| Role | Description |
|---|---|
| Newbie tagger | When you create a new account, you become a newbie tagger. Newbie taggers can create new content and only edit their own content. If newbie taggers wish to edit an existing entry, not created by themselves, they may ask to become a confirmed tagger. |
| Confirmed tagger | A licensed curator of Biii! Confirmed tagger can revert revisions, delete content, edit any content, and publish comments submitted to publication. To parse submitted comments and publish it for confirmed taggers, go to shortcut, validate/edit comments. |
| ALL | Please use the image.sc forum to give us feedback on additional features, report problems in user experience, suggest use cases... |
Entry information standard
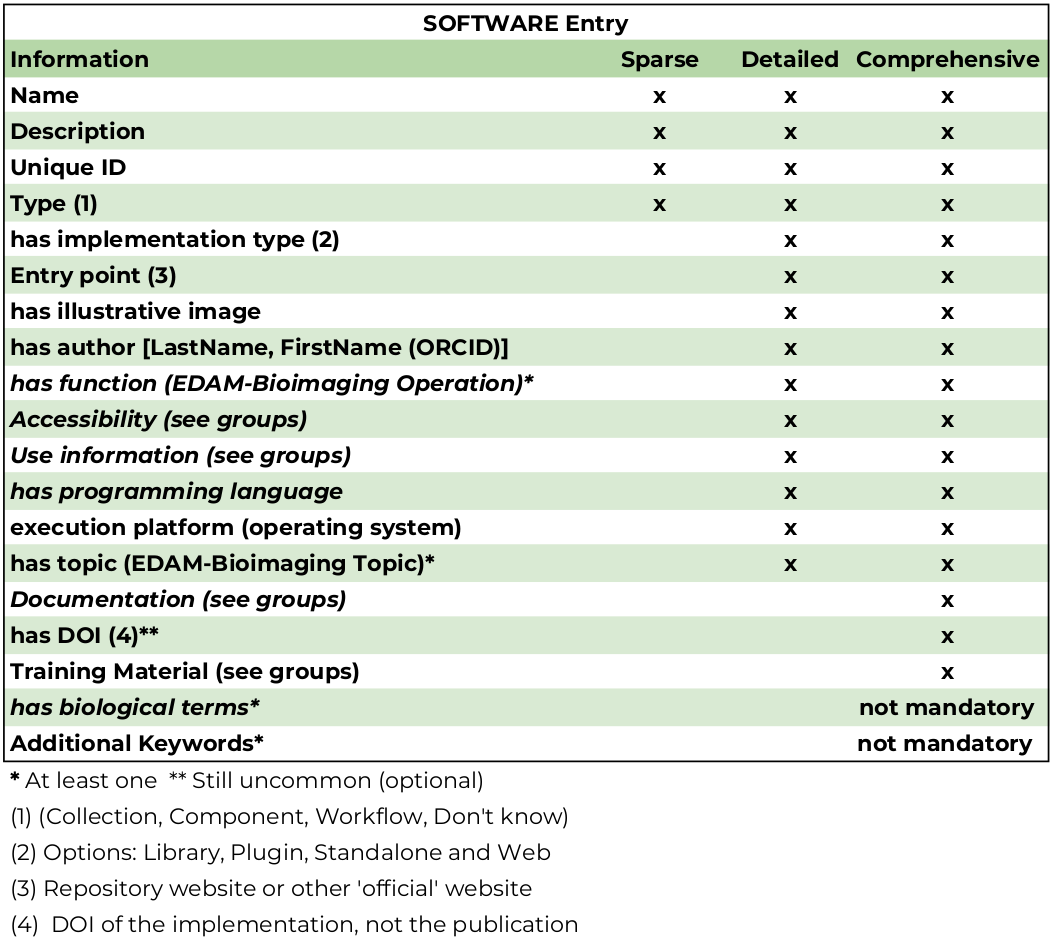
Software entries (not datasets or training materials) will be classified into 3 main tiers relating to its degree of completeness.
The standard described here provides guidelines to support BIII.eu webtool curators to monitor the webtool content and tagging. This standard was adapted from the Tool information standard documentation from ELIXIR bio.tools. Thanks to Jon Ison for referencing this documentation.
This standard comprises a list of entry attributes to be specified for a software entry to be classified in a 5 tier rating of entry completeness and quality in BIII.eu.
BIII.eu includes two ontologies on its framework: Bise core ontology and EDAM-Bioimaging.
In addition, we provide curation guidelines describing how each attribute should be specified to ensure the quality of BIII.eu entries. These guidelines are not limited to the syntactic and semantic constraints defined by EDAM-bioimaging ontology and BISE-core-ontology
The standard provides a basis for monitoring of content and labeling of BIII.eu entries initially by:
-
Entry completeness (3 tiers being "SPARSE", "DETAILED", and "COMPREHENSIVE")
The standard is applied to BIII.eu as follows (condensed information available from the tables):
-
"SPARSE": Minimum information requirement for new entries. A SPARSE entry may be invisible in the webtool (and made it clear to the curator adding the entry in the UI) by default and will only show after additional information is added (similar to what occurs in bio.tools). A sparse entry has a Name, Description, Unique ID (automatically created), and type (collection, component, workflow, don't know). This category may not conform to biii.eu curation guidelines and for curation purposes, such entries must be shown in the list of 'to be curated' of confirmed taggers somehow as 'high priority.
-
"DETAILED": This is the mid-completeness category. An entry with 'Basic details' shall have implementation type (Library, plugin, standalone, and web application), Entry point (URL of the repository or 'official' website), an illustrative image (preferably a screenshot of the UI, or the input/output results, but logo whenever it fits), author ( in the form of
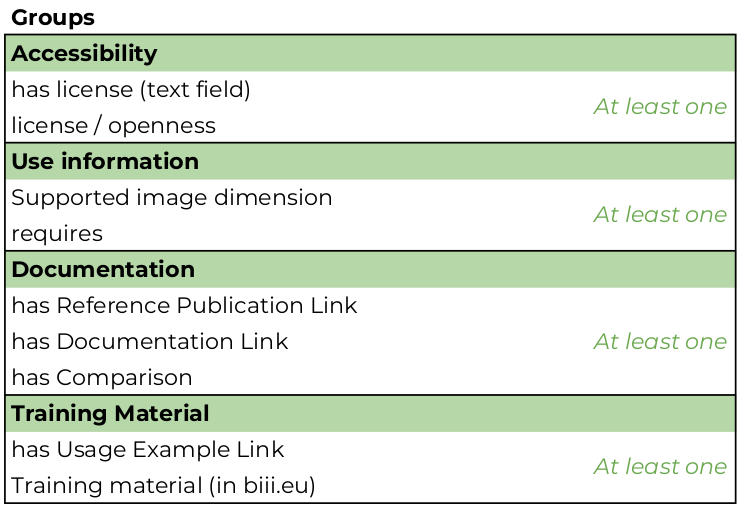
LastName, FirstName (ORCID ID)) and at least one function (EDAM-Bioimaging Operation Class, for example, model-based segmentation or object feature extraction). In this category, there must be at least one attribute from the and Use information groups. In addition, the programming language of the tool is defined as well as its execution platform (Operating System). This category may not conform to biii.eu curation guidelines and for curation purposes, such entries must be shown in the list of 'to be curated' of confirmed taggers without any warning. -
"COMPREHENSIVE": Such entries would provide relevant information to users of all backgrounds and are useful resources and must include at least one attribute from the Documentation group, at least one Topic (EDAM-Bioimaging Topic Class, for example, fluorescence microscopy, Single-molecule localization microscopy, and machine learning), one biological term (which is not a mandatory attribute, but highly recommended) and the DOI of its implementation. Since DOIs of implementations are still not common, this is also an optional attribute (See also: Making your code citable and DOI Registrations for software - DataCite Blog). The additional entry attributes that make an entry comprehensive are Additional keywords (not covered by biological terms or EDAM-Bioimaging) and at least one training material attribute and may have more than just one biological term, EDAM-Bioimaging Operations, and Topics. This category conforms to biii.eu curation guidelines and for curation purposes, such entries must be shown in the list of 'to be curated' of confirmed taggers without any warning.
Guidelines to BIII.eu taggers and curators
BIII.eu is a web-based database that includes bioimage analysis tools, such as Software, Training material, and Datasets. The guidelines presented here will help newbie taggers and confirmed taggers to add and curate software entries into BIII.eu. The software entry of the webtool includes simple components (e.g. Gaussian filter), to image processing libraries, collections of components, and workflows (e.g. single particle tracking).
We ask curators and taggers to include only tools that can be used (e.g we do not seek a publication without the implementation of the code publicly available. Commercial software can be accepted if specified as so and relate to image analysis problems in biology (a.k.a bioimage analysis).
The tools are described using two ontologies, BISE-core-ontology and EDAM-Bioimaging. BISE-core-ontology contains the structure of the description of entries in BIII.eu, not only software, and includes entry properties such as author, reference publication, curator, and so on. EDAM-Bioimaging is used as a source of terms to describe the entry with Bioimaging related vocabulary.
In BIII, a software entry describes a bioimage analysis tool, which can be classified as a component, a collection or a workflow. A component is an implementation of a certain image or data processing / analysis algorithms. Each component alone does not solve a Bioimage Analysis problem. These problems can be addressed by combining such components into workflows. On the other hand, a workflow is a set of components assembled in some specific order to process bioimages and estimate some numerical parameters relevant to the biological system under study. Workflows take image data as input and output either processed images or other types of data (usually numeric values). Workflows can be a combination of components from the same or different software packages. Finally, a collection is software that encapsulates a set of bioimage components and/or workflows, e.g. libraries such as imglib2 and scikit-image or general-purpose software such as Fiji, Icy.
OBSERVATION: These curation guidelines were inspired by biotoolsDocs documentation. Part of it has been adapted or used as-is from the original biotoolsDocs. Thanks to Jon Ison for referencing biotools documentation efforts.
If you wish to suggest changes or additions to this documentation, please raise an issue to begin a discussion.
How and what to curate?
Try to fill as many fields as possible. If one field definition is unclear, report it in the image.sc forum or raise an issue in BIII Github documents repository. Also mind that there is an Entry Information Standard documentation for BIII, where you can check how your entry will be interpreted and curated by confirmed taggers. The entry information standard divides the entry based on its degree of completeness, from 'sparse' to 'comprehensive'.
The keywords "MUST", "MUST NOT", "REQUIRED", "SHALL", "SHALL NOT", "SHOULD", "SHOULD NOT", "RECOMMENDED", "MAY", and "OPTIONAL" in this document are to be interpreted as described in RFC 2119:
-
"MUST", "REQUIRED" or "SHALL" mean that the guideline is an absolute requirement of the specification.
-
"MUST NOT" or "SHALL NOT" mean that the guideline is an absolute prohibition of the specification.
-
"SHOULD" or "RECOMMENDED" mean that there may exist valid reasons in particular circumstances to ignore a particular guideline, but the full implications must be understood and carefully weighed before doing so.
-
"SHOULD NOT" or the phrase "NOT RECOMMENDED" mean that there may exist valid reasons in particular circumstances when acting contrary to the guideline is acceptable or even useful, but the full implications should be understood and the case carefully weighed before doing so.
-
"MAY or "OPTIONAL" means that the guideline is truly optional; you can choose to follow it or not.
Before you start
When you add new content, you will notice if similarly named tools exist. Please check if it is the same tool(s) you wanted to tag, if not, name it differently (example: Erosion (Icy) vs Erosion (MorpholibJ)). The purpose is to have a unique title for each entry. Whenever possible, try to add both the collection and entry by component(s): e.g. MorpholibJ is an ImageJ plugin, and an entry by function (erosion in MorpholibJ, watershed in MorpholibJ, etc.. are all components inside MorpholibJ). The purpose is then to help analysts to find the component they need when constructing a bioimage analysis workflow.
Consider the following before adding a software entry in BIII.eu
1. Are one or more entries required to describe the software?
-
Software with multiple implementations often requires multiple entries. Example: SOAX - has 2 different ways of downloading it: One is the windows standalone executable and the second is the source code, which will require one to compile it in any OS. In that case, the best way is to create two entries: one by location of download since it is actually two different software, and each entry should have only one download page (has location). In addition, one entry would only have Windows as the supported platform, while in the other, any OS.
-
Collections also often require multiple entries. Example: Simple Neurite Tracer (existing entry) is an ImageJ-Fiji plugin which has three workflow options available: 2 semi-automated and 1 fully automated. In this case, one would create one entry for Simple Neurite Tracer (the plugin) as a Collection, then one entry by example workflow, and specify the difference in the title. You will have one plugin, and 3 workflows requiring the plugin Simple Neurite Tracer.
-
tools with multiple interfaces (not described in bise-core-ontology) SHOULD be described by a single entry unless these interfaces provide fundamental functional differences.
-
if in doubt, start a discussion in image.sc forum or contact confirmed taggers or admin.
2. What if the software is already registered?
-
if you are the rightful owner of the entry (i.e. the tool developer or provider of an online service) and you are a confirmed tagger, change the field entry curator to your username. In case you are not a confirmed tagger, ask for a modification of your user status in the BIII.eu on this page.
-
if you are a normal user but wishes to suggest modifications to an existing entry, report it to image.sc forum .
3. Are there version-specific considerations?
-
as a rule, a software entry in BIII.eu SHOULD describe the latest version available at the time of registration and SHOULD be updated, as required, for subsequent releases. Note that revision of your own entries is always available, so if you update to describe a specific version, please state so in the revision log.
-
if a new version has fundamental functional differences it MAY be registered as an entirely new entry. In such cases, follow carefully the guidelines for entry name and version (see Name).
4. Plan how to describe the entry functions by having a look to the EDAM vocabulary, for instance on List of taxonomy terms | BIII.
Attribute Guidelines
The guidelines below are organized into sections as they appear in the create software page in BIII.eu webtool.
Name (Software)
Canonical software name assigned by the tagger, preferably the software developer or service provider, e.g. "Fiji"
1. MUST use name in common use, e.g. in the tool homepage or publication.
2. MUST use short form if available e.g. MaMuT not MaMuT: A Fiji plugin for the annotation of massive, multi-view data.
3. MUST NOT include general or technical terms ("software", "application", "server", "service", "plugin", "app", "add-on" etc.) unless these are part of the common name
4. MUST NOT misappropriate the names of other tools, e.g. there are many Erosion implementations; Calling any of them "Erosion" would be wrong
5. MUST NOT include version information unless this is part of a common name (even if there is still no field for version)
6. SHOULD preserve original capitalisation e.g. MaMuT not mamut.
7. SHOULD follow the naming patterns (see below)
Naming Patterns
For components that are part of a collection, use the pattern {collectionName} toolName For tools that simply wrap or provide an interface to some other tool (NOTE: this is still uncommon in bioimage analysis and was a particular situation in biotools. It needs more discussion), use the pattern
{collectionName} toolName {API|WS}{(providerName)}` *e.g.* `EMBOSS water API (ebi)
where:
-
collectionNameis the name of the library, main software in which the component is present natively or other collection the underlying tool is from (if applicable). -
toolNameis the canonical name of the underlying tool -
use
APIfor Web APIs orWSfor Web services -
providerNameis the name of the institute providing the online service (if applicable)
If in exceptional cases (i.e. when registering, as separate entries, versions of a tool with fundamental differences, substitute for toolName in the pattern above:
toolname versionID` *e.g.* `ilastik 0.5
where versionID is the version number.
Tip: * in case of multiple related entries be consistent, e.g. Open PHACTS and Open PHACTS API * be wary of names that are very long (>25 characters). If shortening the name is necessary, don't truncate it in a way (e.g. within the middle of a word) that would render it meaningless or unintuitive
Description
Textual description of the software, e.g. "The neuTube is a collection of neuron reconstruction tools from fluorescence microscope images. It has an interactive system with a 3D viewer, which can be clicked in 3D and perform neuron tracing automatically and semi-automatically. It can automatically recognize branching points as junctions. Traced neurons can be exported to swc format, which could be imported by various software packages. neuTube has Win and Mac OS standalone executable builds and may also be installed by manual compilation."
example 2: "All-path-pruning 2.0 (APP2) is neuron tracing (fully automated) component of Vaa3D. APP2 prunes an initial reconstruction tree of a neuron’s morphology using a long-segment-first hierarchical procedure instead of the original termini-first-search process in APP. APP2 computes the distance transform of all image voxels directly for a gray-scale image, without the need to binarize the image before invoking the conventional distance transform. APP2 uses a fast-marching algorithm to compute the initial reconstruction trees without pre-computing a large graph. This method allows to trace large images. This method can be used with default parameters or user-defined parameters."
1. MUST provide a concise summary of purpose / function of the tool. we RECOMMEND the description ca. 1-2 short paragraphs.
2. MUST begin with a capital letter and end with a period ('.')
4. SHOULD NOT include any of the following, unless essential to distinguish the tool from other entries:
provenance information e.g. software provider, institute or person name
5. SHOULD NOT describe how good the software is (mentions of applicability are OK)
6. SHOULD NOT include URLs
Author
One or more strings that identify the author(s) of the tool.
1. Each author item MUST correspond to a single individual. In case an individual is not known to be the author, the name of an institution is RECOMMENDED.
2. Each author item MUST follow the following pattern: LastName, FirstName, where LastName MUST NOT be abbreviated and FirstName SHOULD NOT be abbreviated (usually when the first name is long it is RECOMMENDED to use full FirstName abbreviations or partial abbreviations (e.g. SCHOLZ, LEANDRO A. (orcid.org/0000-0002-2411-0429)).
3. SHOULD (if available) include the author's ORCID ID with the following template: LastName, FirstName (orcid.org/xxxx-xxxx-xxxx-xxxx) so they can be contacted more easily (to get the Orcid , google orcid + author name to get it. orcid.org/xxxx-xxxx-xxxx-xxxx).
Illustrative image
An illustrative image that represents the main functionality of the software entry.
1. It SHOULD represent the main software functionality or a screenshot of the UI in use. In cases where a single image cannot show the main functionality of the software entry (usually happens for general-purpose software and libraries) the illustrative image SHOULD be the logo. The software entry will not be promoted on the front page without an illustrative image.
License/Openness
There are only 4 discrete values for this attribute: Commercial, Free and open source, Free but not open source, and I do not know. - Commercial is used when the software needs to be purchased in order to be used. - Free and open source is selected when the source code is available and the software does not need to be purchased in order to be used. - Free but not open source is used when the source code is not available (closed source) but the software is free to be used. - I do not know is used when the License/Openness is not known. This value SHOULD be avoided.
1. If an entry is a Shareware software with different License/Openness values (e.g. a commercial and a free version with fewer features), it SHOULD have both values selected. However, we discourage users to add entries of such type.
Entry curator
A link to a BIII.eu user who is either: (1) the user who added the entry, but who is not the rightful owner of the tool (When the tool is first added), (2) a confirmed tagger, who checked the entry and curated it or (3) the rightful owner of the entry (i.e the tool developer or provider of the online service). When the tool added in BIII.eu, the value of Entry curator will change upon curation (either to the confirmed tagger who curated the tool or the rightful owner of the tool)
Download page
Homepage of the software, from which is possible to download the software or some URL that best serves this purpose, e.g. "http://icy.bioimageanalysis.org/"
1. MUST resolve to a web page from the developer / provider that most specifically provides a downloadable version of the software or has a link to its source code.
2. MUST be restricted to http(s?)://[^\s/$.?#].[^\s]*
3. The link to a Download page that does not work anymore SHOULD be removed and replaced by a new, working link.
TIP: In case a tool lacks its own website, a URL of its code repository is OK. Do not use a general URL such as an institutional homepage.
Reference publication
An url that links to a reference publication that presents the tool.
1. MUST resolve to a web page of a journal article or web page of a preprint server that most specifically links to a reference publication.
2. SHOULD preferably be a DOI link in the form https://doi.org/+DOI (e.g. https://doi.org/10.1371/journal.pbio.1002128). Normal URL links to the reference publication are OK but discouraged.
3. SHOULD preferably resolve to a publication where the tool was first introduced.
4. MAY receive more than one Reference Publication (use Add another item button).
5. The link to the reference publication that does not work anymore SHOULD be removed and replaced by a new, working link.
Documentation
An URL that links to a source of information about the use, installation, and applications of the software. Accepts more than one documentation attribute entry.
1. MUST resolve to a web page from which one can obtain information about how to use the software. For example, a link to a user’s manual (e.g. Neural Circuit Tracer main page, or the link to the User guide itself), a wiki page or other links from which similar information can be obtained (example, Using Fiji page or a readme page with detailed information about the software).
2. From all the options above, it is RECOMMENDED that, if only one Documentation attribute is given, the URL to the documentation resolves to the most used and comprehensive source of information about the software, no matter its type (wiki, pdf file, web page linking to other pages, etc..).
3. MAY receive more than one Documentation link (use Add another item button).
4. The link to a documentation page that does not work anymore SHOULD be removed and replaced by a new, working link.
Has usage example
An URL that links to a usage example, such as a case study document (pdf, web page, video or other types of media), training material (also of any type of media, but preferably existing in BIII.eu), a workflow in which the tool is used (for components).
1. MAY link to an existing node in BIII.eu database (e.g. a workflow, a training material, a dataset).
2. MAY receive more than one usage example link (use Add another item button).
3. The link to a usage example that does not work anymore SHOULD be removed and replaced by a new, working link.
Has comparison
An URL that links to a document, preferably in written media (pdf, slide deck), showing a comparison of the tool against other similar tools that perform the same job or very similar job. Examples: link to a research paper that benchmarks several tools, link to the web page of a Challenge in which the tool is included, reference to other web pages were the comparison is available. BIAFLOWS, the benchmarking webtool from Neubias WG5 could be a source of such attribute. However, we are still discussing how to interact with it.
1. MUST resolve to a web page that shows the results of a comparison of the tool against other similar tools.
2. It is RECOMMENDED that the description (Link text) of the URL indicates the part of the document (a figure, a page or a reference in that document) where the results of the comparison are.
3. MAY link to an existing node in BIII.eu database (e.g. a training material).
4. MAY receive more than one comparison link (use Add another item button).
5. The link to a comparison that does not work anymore SHOULD be removed and replaced by a new, working link.
DOI link ** (DOI of the software)
A single DOI link to the software, related to the correct version of the tool described in the entry. There are many options out there (see this blog post from Datacite), but the most commonly used is Zenodo.
1. MUST be a DOI link in the form https://doi.org/+DOI (e.g. https://doi.org/10.5281/zenodo.30769 ).
Has Training material
A link to an existing training material node in the BIII.eu database.
1. MUST link to an existing training material node in BIII.eu database (e.g http://biii.eu/node/1366).
2. MAY receive more than one training material link (use Add another item button).
The following entry attributes (Has function, Has Topic, Has biological terms) may be considered the most important in BIII.eu. It is with them that the database will be able to connect the tools and make them searchable by bioimage analysts, developers and biologists.
Has function (EDAM-Bioimaging)
Details of a function the tool provides, expressed in concepts from the EDAM-Bioimaging Operation ontology, e.g. image classification and model-based segmentation.
1. MUST correctly specify operations performed by the tool, or (if version indicated), that specific version (s) of the tool.
2. MAY receive more than one Function, especially when the tool has multiple modes of operation.
3. SHOULD describe all the primary operations and SHOULD NOT describe secondary or minor operations. In case there are any questions, start a discussion in image.sc forum or contact confirmed taggers or admin.
Has Topic (EDAM-Bioimaging)
General scientific domain the tool serves or other general categories (EDAM Bioimaging Topic), e.g. Tissue image analysis, Microscopy, Machine Learning.
1. MUST specify the MOST IMPORTANT and relevant scientific topics, although we RECOMMEND referring at least to the single most important scientific topic.
2. MUST correctly specify Topics the tool relates to, or (if version indicated), those specific version(s) of the tool.
3. MAY receive more than one Topic. (use Add another item button)
4. SHOULD NOT exhaustively specify all the topics of secondary relevance. Include Topic(s) that include the tool into the pool related tools (with the same Topic). And, if applicable, include one or more Topics that distinguish the tool from the others in the pool. (see discussion in EDAM-Bioimaging.
Has biological terms
1. MUST specify the most important and relevant biological terms.
2. MAY receive more than one biological term (use Add another item button).
Additional keywords
A string in which the user may add keywords to the entry in case she/he did not find existing keywords in EDAM-Bioimaging functions or topics. This field is important to support further improvements and discussions on new versions of EDAM-Bioimaging or modifications in BIII.eu.
1. MUST be a concise keyword and comprise of the most commonly used keyword that relates to the intended theme/subject (to the best of the user's knowledge, for we do not expect the user to know the best term, which is also not always agreed upon by the scientific community).
2. MAY receive more than one keyword (use Add another item button).
Requires
A link to an existing software node in BIII.eu to show in which platform it can be run or the dependencies of the tool. For example tool 3D intensity profile requires ImageJ to be run. On the other hand, DeconvolutionLab2 ImageJ plugin, not only requires ImageJ but also other libraries, such as (under construction).
Execution platform
A discrete attribute that defines in which main execution platforms the tool can be used. There are only 4 values for this attribute: Linux, Mac, Windows, Unsure.
Implementation type
A discrete attribute that defines the type of implementation of the tool. A more detailed description of the discussion of implementation types is in "Workflows and Components of Bioimage Analysis: The NEUBIAS Concept" . The 4 values for this attribute are
Collection, Component, Workflow and I do not know
1. Component is an implementation of an image or data analysis/processing algorithm that may be used as a part of an image analysis workflow. A component alone does not solve a Bioimage Analysis problem.
2. Workflow is a set of components assembled in some specific order to process bioimages and estimate some numerical parameters relevant to the biological system under study. Workflows take image data as input and output either processed images or other types of data (usually numeric values). Workflows can be a combination of components from the same or different software.
3. Collection is a software comprising a group of Components or Workflows. Collections are often image analysis platforms or libraries, e.g. scikit-image library and ImageJ platform.
4. I do not know is a tool whose type is unknown. Ideally, the user adding the entry SHOULD identify the tool type prior to adding it to BIII.eu.
License
Software or data usage license, e.g. "GPL-3.0"
1. MUST accurately describe the license used.
2. SHOULD use "Proprietary" in cases where the software is under license whereby it can be obtained from the provider (e.g. for money), and then owned, i.e. definitely not an open-source or free software license.
3. SHOULD use "Unlicensed" for software that is not licensed and is not "Proprietary".
4. SHOULD either use "Other" or the License name (if known) if the software is available under an uncommon license not listed below and which is not "Proprietary".
Most Common Licenses (if you know of an important one to add feel free to suggest):
-
Apache License 2.0
-
BSD 3-Clause "New" or "Revised" license
-
BSD 2-Clause "Simplified" or "FreeBSD" license
-
GNU General Public License (GPL)
-
GNU Library or "Lesser" General Public License (LGPL)
-
MIT license
Has programming language
Comprises both programming language used for the implementation of the entry and the programming languages supported by the entry. We still do not have a controlled vocabulary, so if you type a new language, which was not previously added in BIII.eu, it will create a new node with that name.
is compatible with
A link to an existing software node in BIII.eu for which the tool was not originally developed, but that can be called from.
Supported image dimension
A discrete attribute value that defines with which image dimensions the tool can be used. The four discrete values are 2D, 3D, Multi-channel, and time-series. OBSERVATION: Some may understand there is an overlap with these values, for a 2D RGB image would also be a Multi-channel image and a 3D image could be a 2D + time-series image, etc.
Interaction level
A discrete attribute value that defines the interaction level between the user and the tool. There are four discrete values, which are:
1. Automated a tool that returns the output with a single command call (selection in a GUI) that may or may not accept the definition of parameters.
2.Manual a tool constructed in a way such that it uses a user interface, most commonly a Graphic User Interface (GUI) to help users perform manual image analysis tasks. ImageJ multi-point tool.
3.Semi-automateda tool that requires multiple calls or user interactions in order to deliver the output. For example, tracing filaments individually until all filaments of an image are traced or identifying central points of cells in order to segment them. The interaction may occur prior to the execution of the tool or several times while the tool is being used (e.g. Simple Neurite Tracer)
4.I do not know a tool that does not have a known interaction level. Ideally, the tagger adding the entry to BIII.eu SHOULD identify the interaction level prior to adding the entry.
Comments
Comments are a valuable part of BIII.eu. The comments allow users to talk more specifically about a certain tool.
Users may leave comments on each page of BIII.eu entries. The content of the comments may include an opinion about the tool performance (how good the tool does what was said to do), use cases, or an update on the current status of the tool (deprecated, legacy, etc..). Users MUST be polite and avoid rude, violent comments.
The information contained here is available in NEUBIAS GitHub repository under bise-documents
- 4505 views